
Computational biology, cancer immunotherapy, and structural bioinformatics
My research group uses structural bioinformatics methods, such as molecular modeling, molecular docking and molecular dynamics, to investigate protein-ligand interactions with relevant biomedical applications. For instance, during the COVID-19 pandemic, I have used molecular dynamics simulations to study a peptide-based inhibitor of the complement system with potential use for COVID-19 treatment, and I have used Markov-State Modeling to study molecular interactions involved in the recognition of a particular SARS-CoV peptide by the immune system. Since we are constantly pushing the limits of what can be done with available tools, my group is also actively adapting and developing new computational methods to address specific biological problems. For instance, we recently implemented a webserver which accounts for the role of receptor flexibility when predicting the binding of potential inhibitors for SARS-CoV-2 proteins.
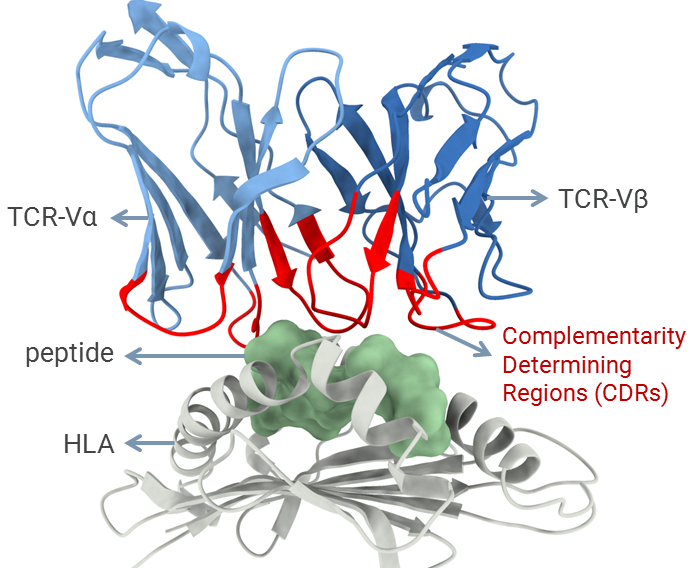
In addition to collaborative projects involving broader biomedical applications, my lab has a particular focus on studying the mechanisms involved in cellular immunity. This type of adaptive immunity is mediated by T-cell lymphocytes and is key for immunological responses targeting both viruses and cancer cells. T-cells can recognize pieces of proteins (i.e., peptides) displayed at the surface of other cells by a family of receptors known as human leukocyte antigens (HLAs). The recognition of peptide-HLA complexes is mediated by the complementarity determining regions (CDRs) of the T-cell receptor (TCR), representing a central step for the activation of T-cells and the development of cellular immunity. Understanding the molecular features driving the affinity and specificity of the TCR/pHLA interaction can create new opportunities for biomedical applications across different fields, including antiviral vaccine development, cancer immunotherapy, and the treatment of autoimmune diseases. Over the past decade, I have developed several tools to enable the computational modeling and structural analysis of peptide-HLA complexes. Now I want to combine these computational methods with data from high throughput molecular biology and proteomics approaches, to improve the safety and the efficacy of future T-cell-based immunotherapies.
For more on our research projects, please check the Research and Publications tabs.
New computational lab space at the Center for Nuclear Receptors and Cell Signaling
I have a new lab space inside the Center for Nuclear Receptor and Cell Signaling (CNRSC). We are on the process of purchasing new high performance computers that will be for exclusive use of researchers and students in my group. In addition, my group will have access to shared facilities of the CNRCS, and core facilities of the Department of Biology and Biochemistry. These include the University of Houston Sequencing and Editing Core (UH-SNEC) and the Research Computing Data Core. Our Department is part of the Gulf Coast Consortia for Quantitative Biomedical Sciences (GCC), which offers unparalleled networking and training opportunities, including fellowship programs through the Keck Center.
Information for prospective PhD students
Our department offers PhDs in both Biology and Biochemistry, with a commitment for at least 5 years of financial support (TA/RA, plus a partial tuition fellowship). In addition to the core and elective courses offered by our department, Biochemistry students can take elective coursework within the departments of Chemistry, Physics, or Math; our students can also take courses offered by the Graduate School of Biomedical Sciences at UT Health.
Our Biochemistry Division includes 14 faculty across a broad range of research areas, including structural biology, gene regulation through chromatin modifications, cancer metabolism, genomics, microbial gene regulation, RNA structure/function, structural biochemistry underlying immune recognition and function. Ongoing collaborations within this group include faculty in the departments of Chemistry, Chemical Engineering, Biomedical Engineering, and Pharmacological and Pharmaceutical Sciences.
Our students can also participate in a range of professional development opportunities offered through our department (workshops, panels, etc), as well as similar events offered through the Texas Medical Center. Some of our students have also completed part-time biotech internships through a local network.
For additional information, please check the admissions webpage or contact our Graduate Program Advisor, Ms. Rosezelia Jackson (biograd@central.uh.edu, Tel: (713) 743-2633).
